This blog post was written by Taylor Smith, the 2019 Kathryn Turner Diversity and Technology Intern in the Smithsonian Libraries’ Web Services Department. At the time of her internship, Taylor was an undergraduate Computer Science student at Bowie State University. Her work in the summer of 2019 consisted of developing and coding a method for identifying article metadata in The Avicultural Magazine, a leading journal for the keeping of non-domesticated birds in captivity.
As a biology major with an interest in computer science, I had a curiosity for wildlife and a newfound love for coding. I kept the two in mind when searching for internships, and luckily for me, I was led to the Kathryn Turner Diversity in Technology Internship for the summer of 2019. When I saw that the internship would focus on working with zoo articles relating to botany and wildlife, I knew this was perfect for me.
I had never held an internship before, let alone one that involves coding (which I had started learning that year). I had no idea what to expect when coming into this internship, but I learned a lot more than I could have imagined. Throughout this internship, I learned what metadata was and why it was so important. I learned why having digitized articles available online was so crucial. I also learned that making information accessible took a lot more work than anyone would think.
In my first week, I was introduced to the Biodiversity Heritage Library (BHL), an online digital library designed to make biodiversity literature available to the public. In this library, I was specifically working with The Avicultural Magazine. This was a journal created by the Avicultural Society in 1894 with the purpose of spreading information, advice, and updates on non-domesticated birds. The volumes are digitized by Smithsonian Libraries and Archives and processed through optical character recognition (OCR) for the convenience of zookeepers and other zoo curators. The only problem with this is that it takes scrolling through endless pages of articles to find the specific item you’re looking for. My job was to create code that finds metadata for these articles to make them much more accessible and citable.
Below is an example of a page with the beginning of an article.

At first, I had to write code that would open up the directory of all the articles, open up one file at a time, and look for titles, page numbers, authors, etc. I set to work, but it was not long before we found that Penn State University and the National University of Singapore actually had a project named ParsCit that went through the files and searched for said data. The results are placed into an XML file, which was helpful to the process but not exactly as we needed.
My job then became loading and parsing the XML files using C++. The task was initially daunting: I had no idea what OCR was, what parsing meant, or what an XML file was. But with the help of Joel Richard, my supervisor and head of Web Services, it came a little more smoothly than I expected.
Joel really helped me take the next step in applying myself and taught me about regular expressions, parsing, XML files, and helped me with any trip-ups I had along the way. He also helped re-run the OCR for better material to work with, as well as pre-process the XML files so the data was more consistent and easier to work with.
After receiving the XML files I had to parse them. Parsing is essentially going through the tags of an XML file and picking out specific types of information. The process was not as simple as finding parts of the text marked as “author” or “title”, or even tags marked as such, but rather required me to take a look at the patterns occurring in the file. So, in my case, if I wanted the title, I would most often go to a <sectionHeader> tag since that’s where the Penn State code placed the titles it caught.
I checked the places I knew most of the titles were held (some were not caught at all and were in the middle of body text, which we couldn’t catch) and then looked for uppercase letters since all the titles were capitalized. Then, I checked if there was a “By” in either the <sectionHeader> tag or in the following <bodyText>tag. If nothing was found, then it wasn’t deemed an article. For authors, if there was an article found, then the “By” search would lead to an author every time. The next picture is an example of the OCR from one of the articles. You can see in this particular one the title is in the section header tag, but the author is in the body text tag.
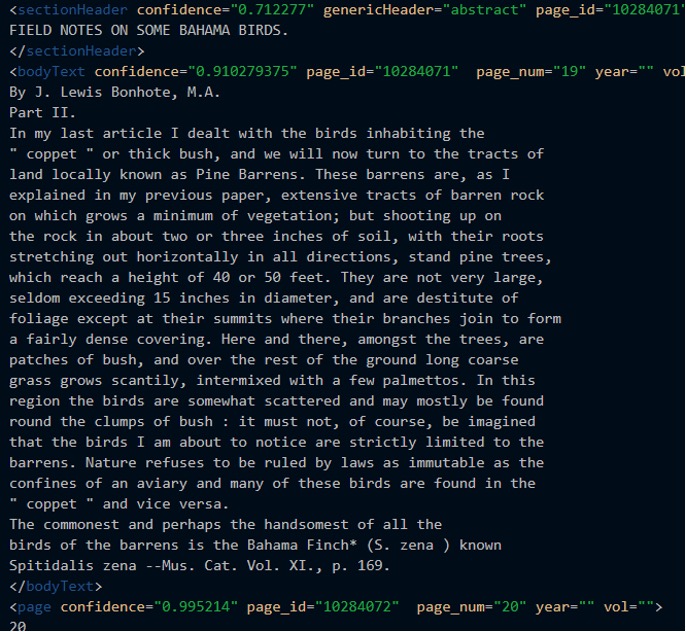
For Page Numbers and Page IDs at BHL, I looked into the attribute of a tag. In the screenshot, you can see page_id and page_num which are attributes of <bodyText>. I pulled the information from those tags and stored them as is.
Since the OCR is imperfect, sometimes the “B” in “By” was interpreted as maybe an “E” or maybe the “V” in “Vol” was found as a “Y”. I did some very specific checking so that for as many cases as possible, as long as the very specific conditions were met, the information was found. We used regular expressions which looked for certain arrangements of these similar characters to determine the author, volume, date, and similar data.
After finding and storing the data, much of the remaining work was cleaning up what I had. That meant taking off any extra spaces, periods, capitalizing the first character of each word, and making the rest lowercase, or maybe refining the results even more. This was to make it easier for the humans who correct the results afterward. The code is effective, but not perfect. When the human cleanup was complete, the articles were imported into the Biodiversity Heritage Library.
While interning, I was able to go on many tours including the Smithsonian Libraries Research Annex collections, the Joseph F. Cullman Library 3rd Library of Natural History, and behind the scenes of the National Zoological Park. From viewing James Smithson’s books at the Cullman to watching the process of restoring aged and delicate books in the Book Conservation Lab to learning about mole rats, each tour showed a different but equally fascinating aspect of the libraries, as well as their involvement with the Zoo. I thoroughly enjoyed seeing the older objects, as it was very cool to see things that have held importance for such a long time and how times have changed.
The Zoo tour was led by branch librarian and supervisor Stephen Cox. Both Stephen and Jackie Chapman, Head of the Digital Library and Digitization, were extremely helpful, generous, and informative throughout the internship. They even helped me and intern Katerina Ozment create a poster to present at the Association of Zoos and Aquariums symposium! Katerina’s internship was focused on manually collecting and analyzing metadata from a related journal, the Animal Keepers’ Forum.
Kathryn Turner, the sponsor of my internship, is an inspiring woman I had the privilege to meet and share my story with her. It was motivating to see another woman with a very similar background to mine rise up in the STEM world and conquer it. I’m so grateful that I had the opportunity to intern and learn at the Smithsonian Libraries (now Smithsonian Libraries and Archives) that summer. I not only was able to expand my knowledge in my field but was also able to meet very intriguing people and see how things work behind the scenes.
Editor’s Note: Since the writing of this post, the National University of Singapore has developed and released a neural net version of their ParseCit software. This greatly improves the potential effectiveness of future computer-based automatic identification of articles building upon Taylor’s groundwork.

Be First to Comment